ChimeraX Recipes
Search AlphaFold database for transmembrane cysteines
This is an example of searching the C elegans proteome for transmembrane proteins that have at least two nearby cysteines in the membrane. This uses AlphaFold database predicted structures and UniProt annotations to identify transmembrane residues. It searches 19827 UniProt entries for C elegans, finds 5756 with annotated transmembrane regions, and then finds 783 proteins with two cysteines in the transmembrane region closer than 5 Angstroms to each other (SG to SG atom distance). The search takes 36 seconds on a Mac laptop, so it is feasible to try many variations of this kind of structural feature search in a short time.
UniProt sequence annotations
In order to make the search fast we download in advance the UniProt annotations for all C elegans proteins as a single XML file (193 Mbytes). The C elegans proteome is described here
https://www.uniprot.org/proteomes/UP000001940
and the XML file is here
https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/reference_proteomes/Eukaryota/UP000001940/UP000001940_6239.xml.gz
AlphaFold database structures
We also download all of the AlphaFold database structures for C elegans in advance (2.6 Gbytes) described here
https://alphafold.com/download
and the file is here
https://ftp.ebi.ac.uk/pub/databases/alphafold/latest/UP000001940_6239_CAEEL_v4.tar
We put the structures in a directory called alphafold_models and uncompress them
mkdir alphafold_models
cd alphafold_models
tar xf ~/Downloads/UP000001940_6239_CAEEL_v4.tar --include '*.cif.gz'
gunzip *.gz
Running the search in ChimeraX
Opening the Python script cyssearch.py in ChimeraX 1.7 then does the search and prints the results to the ChimeraX log. The script uses standard Python capabilities to read the UniProt annotation XML file, and uses ChimeraX capabilities to read the AlphaFold structures to measure distances between cysteines.
Use ChimeraX commands to change to the directory with the UniProt XML file, Python script and alphafold_models directory, then open the script in ChimeraX.
cd ~/af_search
open cyssearch.py
The following output is logged listing the UniProt ids with residue numbers for pairs of close cysteines. At the end of the output is a list of structures for which there was no AlphaFold model (too new 2022 or too large >1400 amino acids for AlphaFold database).
19827 UniProt entries
5756 entries with annotated transmembrane regions
4518 entries with 1 or more transmembrane cysteines
783 with two cysteines closer than 5A, at least 1 being transmembrane
O16978 209,260
O17230 137,210
O17956 93,97 76,84
O17960 47,91
O18083 17,306 99,153
Q18595 123,468
Q19061 85,93
Q65XS8 243,302
Q9NA41 149,204
A0A067XG43 63,259
A4UVM0 150,216
Q86D06 138,180
Q8WQB6 37,309
E5QCI3 177,217
O16880 634,638
P34315 17,388
Q19975 47,55
Q93655 280,531
Q966F7 18,189
Q9GYL0 145,165
O45767 162,181 33,72
Q9U3P2 1103,1107
G5ED82 141,145 254,323
O16494 97,153
O45971 54,107
Q7YWW4 37,41
Q9GYP2 75,77 72,75 71,83 77,78 71,73 72,78 71,86 73,83 72,77
...
No alphafold model for 44 entries with transmembrane regions:
A0A8S4SQM5 502
Q9UB28 4450
A0A8D9MJW7 973
A0A8D9J776 335
...
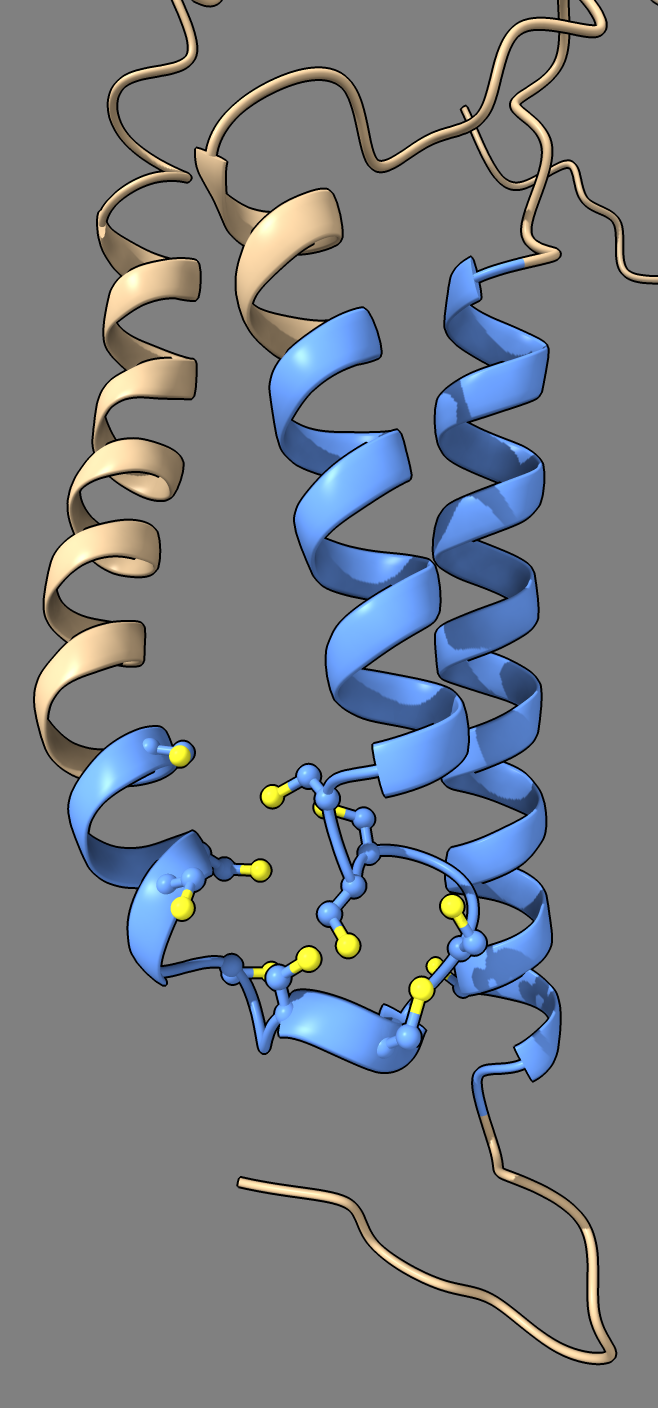
Example structure
Here is an image of the uncharacterized protein Q9GYP2 with a cluster 11 closely spaced cysteines in or near transmembrane helices.
Python script
Here is the ChimeraX Python script that does the search cyssearch.py
# Dengke Ma wants to find all C elegans proteins with pairs of close cysteines in transmembrane regions.
# Can use UniProt to identify transmembrane residues, then use AlphaFold database predicted structures
# to see if there are close cysteines.
def find_uniprot_transmembrane_cysteines(uniprot_xml_path, namespace = '{http://uniprot.org/uniprot}'):
import xml.etree.ElementTree as ET
tree = ET.parse(uniprot_xml_path)
tm = []
for child in tree.getroot():
if child.tag == namespace + 'entry':
rr = transmembrane_residue_ranges(child, namespace)
uniprot_id = child.find(namespace + 'accession').text
seq = child.find(namespace + 'sequence').text
cys_count = ''.join(seq[b-1:e] for b,e in rr).count('C')
tm.append((uniprot_id, cys_count, len(seq), rr))
return tm
def transmembrane_residue_ranges(protein_xml_entry, namespace):
ranges = []
for feature in protein_xml_entry.iter(namespace + 'feature'):
fattrib = feature.attrib
if 'type' in fattrib and fattrib['type'] == 'transmembrane region':
for loc in feature.iter(namespace + 'location'):
b,e = loc.find(namespace + 'begin'), loc.find(namespace + 'end')
if b is not None and e is not None:
if 'position' in b.attrib and 'position' in e.attrib:
r = (int(b.attrib['position']), int(e.attrib['position']))
ranges.append(r)
return ranges
def close_cysteines(structure, membrane_residue_ranges, max_distance = 5):
cys_res = [r for r in structure.residues if r.name == 'CYS']
cys_xyz = [(r.number, r.find_atom('SG').coord) for r in cys_res]
mb_res_nums = set()
for b,e in membrane_residue_ranges:
for rnum in range(b,e+1):
mb_res_nums.add(rnum)
mb_cys = [r for r in cys_res if r.number in mb_res_nums]
mb_xyz = [(r.number, r.find_atom('SG').coord) for r in mb_cys]
close_pairs = set()
from chimerax.geometry import distance
for rnum, xyz in mb_xyz:
for rnum2, xyz2 in cys_xyz:
if rnum2 != rnum and distance(xyz, xyz2) <= max_distance:
pair = (rnum, rnum2) if rnum < rnum2 else (rnum2, rnum)
close_pairs.add(pair)
return list(close_pairs)
def check_for_close_cysteines(session, ulist, alphafold_dir, max_distance):
found = []
missing = []
for uniprot_id, ncys, seq_len, tm_res_ranges in ulist:
if ncys == 0:
continue
m = alphafold_database_model(session, uniprot_id, alphafold_dir)
if m is None:
missing.append((uniprot_id, seq_len))
continue
close_pairs = close_cysteines(m, tm_res_ranges, max_distance)
if close_pairs:
found.append((uniprot_id, close_pairs))
m.delete()
return found, missing
def alphafold_database_model(session, uniprot_id, alphafold_dir):
filename = f'AF-{uniprot_id}-F1-model_v4.cif'
from os.path import join, exists
path = join(alphafold_dir, filename)
if not exists(path):
return None
from chimerax.mmcif import open_mmcif
s, msg = open_mmcif(session, path)
return s[0]
uniprot_xml_path = 'UP000001940_6239.xml'
alphafold_dir = 'alphafold_models'
max_distance = 5
ulist = find_uniprot_transmembrane_cysteines(uniprot_xml_path)
uclose, missing = check_for_close_cysteines(session, ulist, alphafold_dir, max_distance)
ntm = len([uniprot_id for uniprot_id, ncys, seq_len, tm_res_ranges in ulist if tm_res_ranges])
ntmc = len([uniprot_id for uniprot_id, ncys, seq_len, tm_res_ranges in ulist if ncys > 0])
print(f'{len(ulist)} UniProt entries')
print(f'{ntm} entries with annotated transmembrane regions')
print(f'{ntmc} entries with 1 or more transmembrane cysteines')
print(f'{len(uclose)} with two cysteines closer than {max_distance}A, at least 1 being transmembrane')
entries = []
for uniprot_id, res_pairs in uclose:
rpairs = ' '.join(f'{r1},{r2}' for r1,r2 in res_pairs)
entries.append(f'{uniprot_id} {rpairs}')
print()
print('\n'.join(entries))
print()
me = '\n'.join(f'{uniprot_id} {seq_length}' for uniprot_id, seq_length in missing)
print(f'No alphafold model for {len(missing)} entries with transmembrane regions:\n{me}')
Modifications
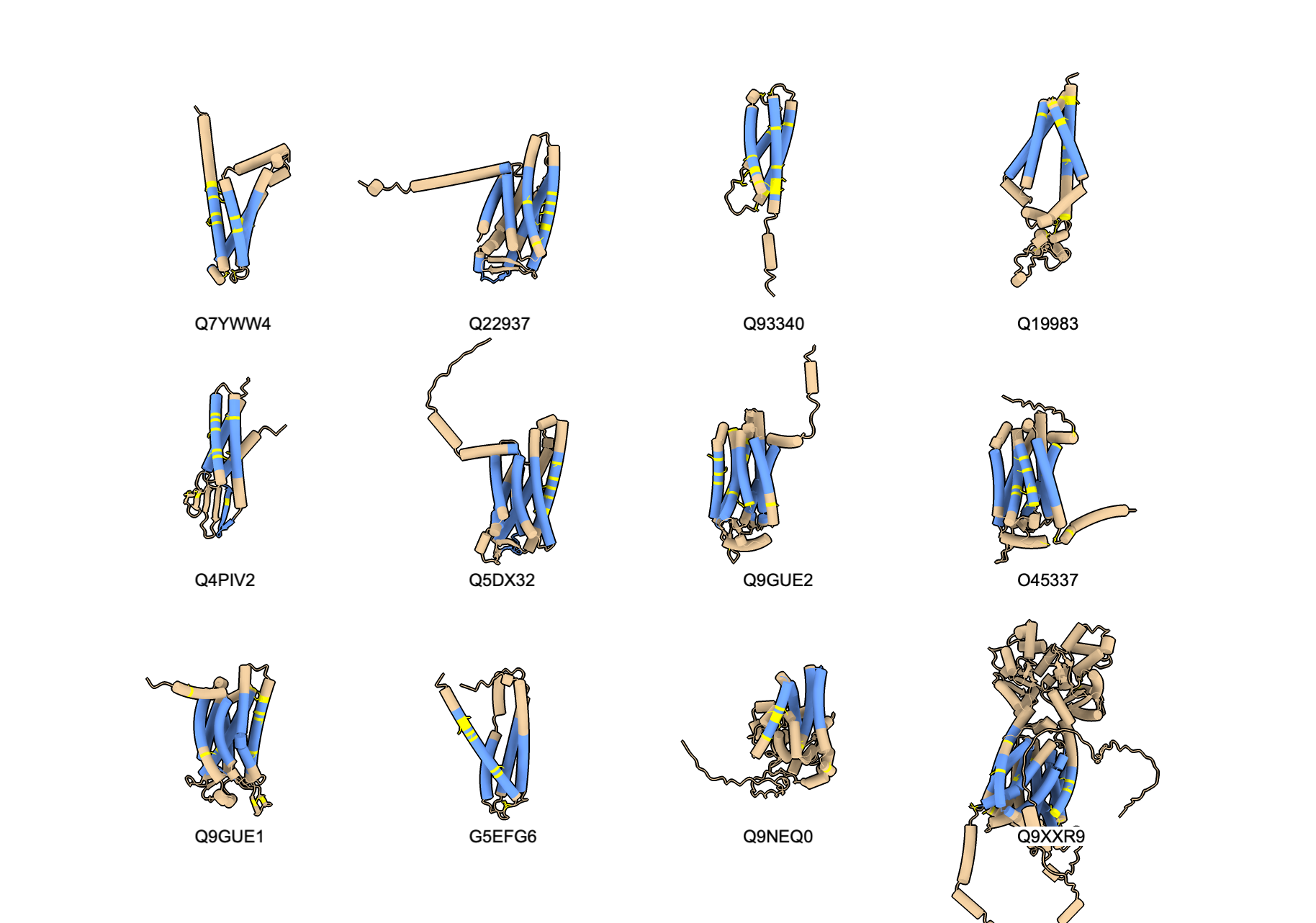
The script can easily be modified to instead find all C elegans proteins with at least 4 transmembrane helices where one of those helices has at least 5 cysteines. And we can load those structures with the transmembrane helices selected. Here is the script cyssearch_v2.py that does that, and the 12 structures found are listed and shown below.
19827 UniProt entries
5756 entries with annotated transmembrane regions
4518 entries with 1 or more transmembrane cysteines
12 UniProt entries with 5 cysteines in one transmembrane helix and at least 4 transmembrane helices
# UniProt ID, protein name, number of transmembrane segments, maximum single segment cysteines)
Q7YWW4,Conserved plasma membrane protein,4,5
Q22937,Serpentine receptor class gamma,7,5
Q93340,Candidate tumor suppressor protein,4,7
Q19983,,4,5
Q4PIV2,Claudin domain-containing protein 1,4,5
Q5DX32,,7,6
Q9GUE2,Seven TM Receptor,7,6
O45337,Seven TM Receptor,7,5
Q9GUE1,Seven TM Receptor,7,5
G5EFG6,Small integral membrane protein,4,8
Q9NEQ0,,4,5
Q9XXR9,SSD domain-containing protein,11,5
More modifications
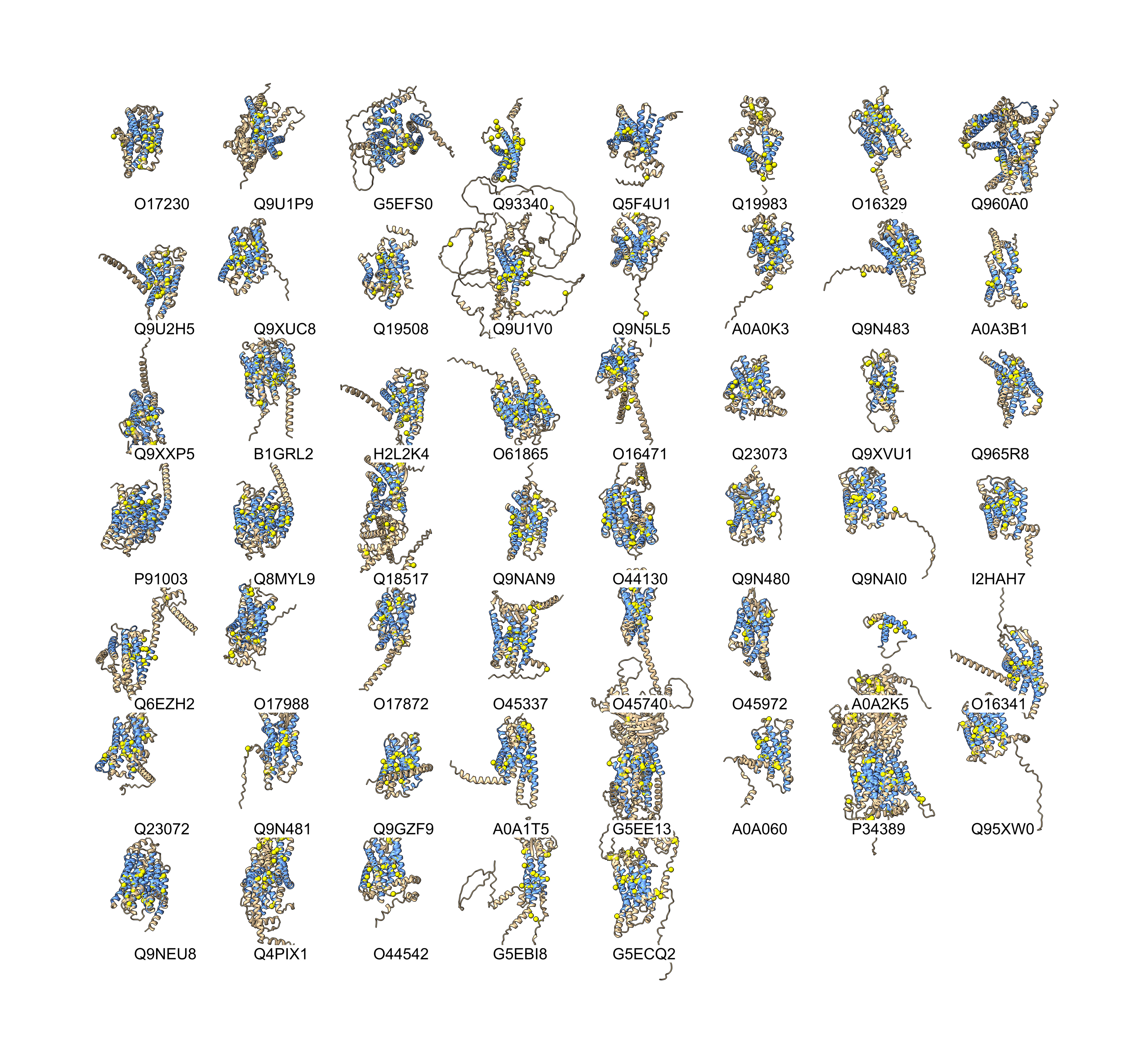
Denke Ma suggested filtering to proteins with at least 4 transmembrane helices and looking for pairs of cysteines separated by 0, 1, or 2 residues (CC, CxC, CxxC) in each helix and requiring two helices to have pairs with distance at most 10 Angstroms from a cysteine SG atom in one helix to the other. That gave 53 proteins results_v3.txt using script cyssearch_v3.py.
Tom Goddard, January 19, 2024