ChimeraX Recipes
Set atom radius using bfactor
Note that beginning with the ChimeraX 1.8 daily build, there is now an “size byattribute” command that provides the same kind of functionality as this recipe. The commands to exactly duplicate the recipe example would be:
open 7w91
size byattr bfactor 20:.6 300:9
though simply:
open 7w91
size byattr bfactor
produces a very reasonable result.
Eric Pettersen, November 22, 2023
Previous recipe for reference
Here is Python code defining a command “atomsize” that sets the radius of atom spheres to be proportional to an attributre such as bfactor. Zhiyuan Liu asked about this for showing gene expression on genome structure models in ChimeraX. Here is an example that shows atom radii proportional to bfactor.
open atomsize.py
open 7w91
atomsize #1 attribute bfactor scale 0.03
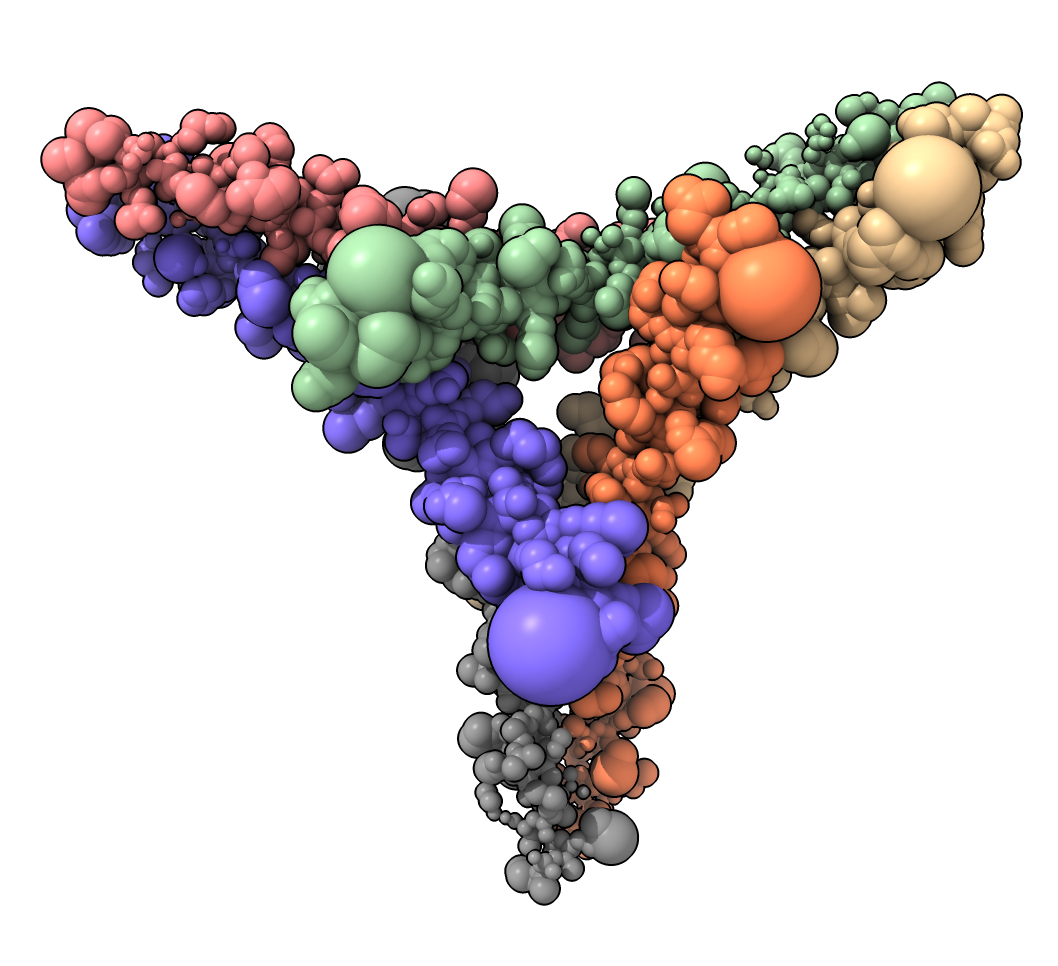
and here is the atomsize.py code:
def atomsize(session, atoms, attribute = 'bfactor', scale = 0.02, offset = 0.0):
for a in atoms:
value = getattr(a, attribute, None)
if value is None:
value = getattr(a.residue, attribute, None) # Try residue attribute
if value is not None:
a.radius = offset + scale * value
a.draw_mode = a.SPHERE_STYLE
def register_command(logger):
from chimerax.core.commands import register, CmdDesc, StringArg, FloatArg
from chimerax.atomic import AtomsArg
desc = CmdDesc(required = [('atoms', AtomsArg)],
keyword = [('attribute', StringArg),
('scale', FloatArg),
('offset', FloatArg),
],
synopsis='Set atom sizes proportional to an attribute value')
register('atomsize', desc, atomsize, logger=logger)
register_command(session.logger)
Tom Goddard, July 24, 2023