ChimeraX Recipes
Read CASTP pockets file
Our older program Chimera can read CASTP web server molecule pockets files (.poc) and display surfaces for the pockets. That has not yet been implemented in ChimeraX. But the file format is easy to read. Here we make a command that reads a CASTP pocket file and define names in ChimeraX pocket1, pocket2, … that specify the atoms lining each pocket. Opening the python code registers the readcastp command
open read_castp.py
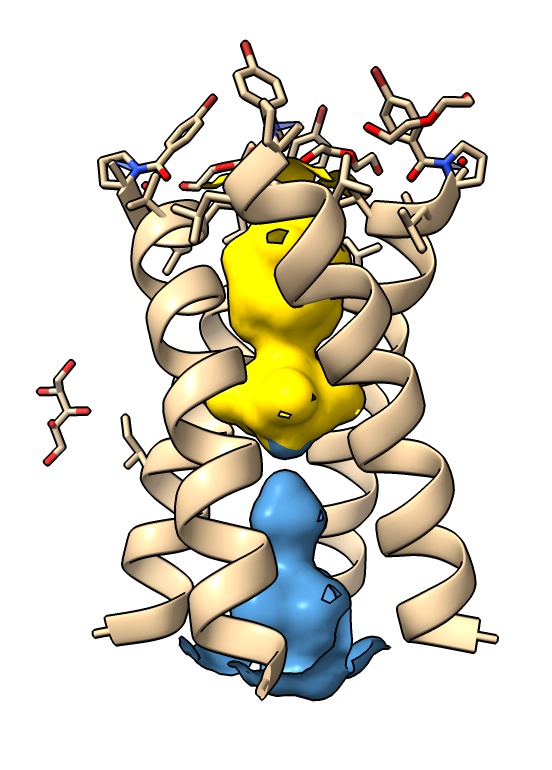
then use the command on an atomic model, here the influenza M2 ion channel,
open 3lbw
readcastp ~/Downloads/3lbw/3lbw.poc
and show and color two pockets
surface pocket1 enclose protein visiblePatches 1
color pocket1 gold surface
surface pocket2 enclose protein visiblePatches 1 replace false
color pocket2 steelblue surface
The .poc file is just a PDB file with the pocket number for each atom at the end
ATOM 165 NH2 ARG A 45 -9.395 -35.779 0.675 1.00 32.68 2 POC
ATOM 194 O VAL B 27 -12.428 -9.493 5.152 1.00 13.76 1 POC
ATOM 196 CG1 VAL B 27 -11.788 -7.554 2.677 1.00 14.96 1 POC
...
The script contructs the ChimeraX atom specifier from chain identifier, residue number and atom name. For example, the above lines are for atoms /A:45@NH2, /B:27@O, /B:27@CG1 in ChimeraX syntax belonging to pockets 2, 1 and 1. It then defines in the code the name pocket1 using command “name pocket1 /B:27@O/B:27@CG1…”.
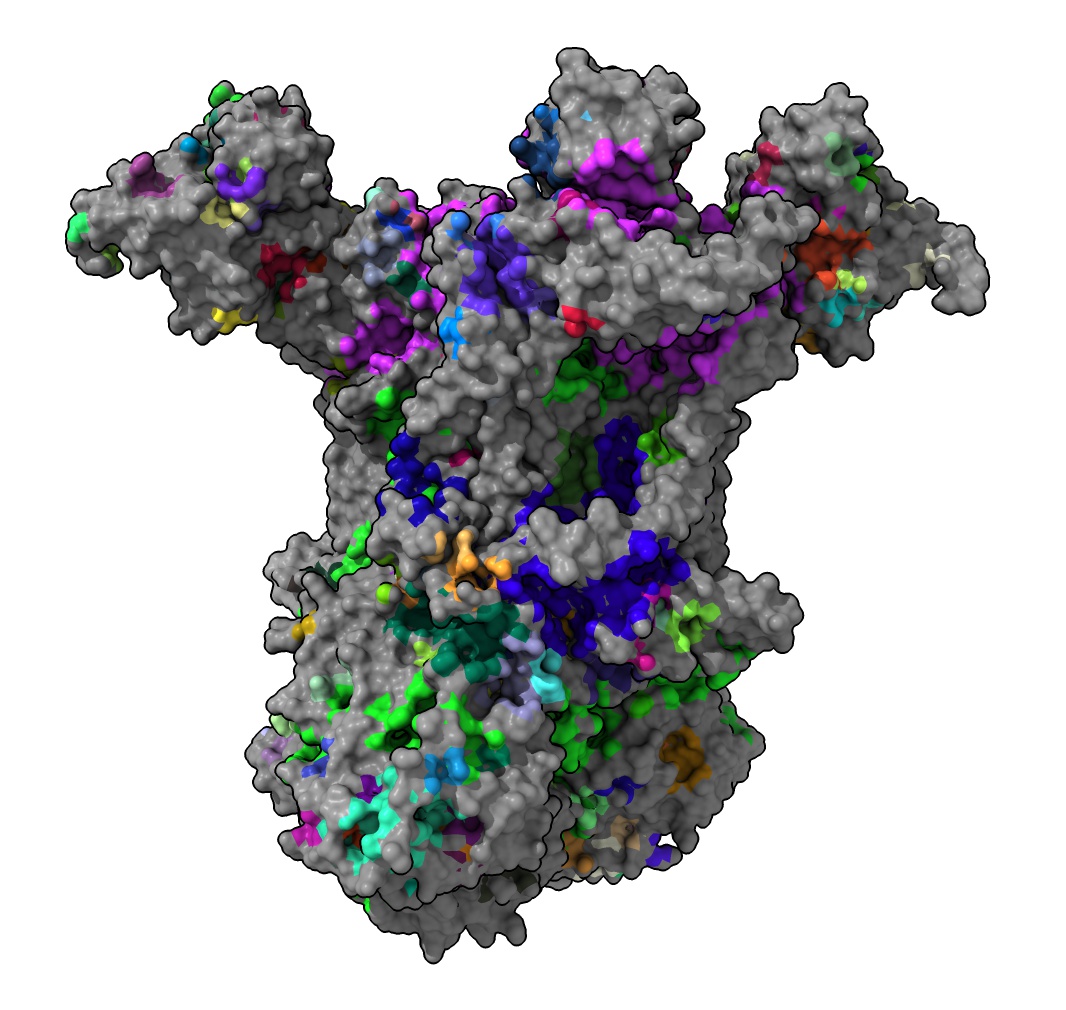
Yaikhomba Mutum asked on the ChimeraX mailing list about looking at pockets of the large complex III electron transfer structure PDB 3CX5. Chimera is unable to compute the surfaces for that large structure but ChimeraX has reliable surface calculation. Here are the 509 pockets colored randomly using
open 3cx5
surface
color gray surface
readcastp ~/Downloads/3cx5/3cx5.poc color true
This takes a few minutes because the command parsing for the couple pockets with thousands of atoms is slow.
Here is the read_castp.py code:
def pocket_atom_specifiers(poc_file_path):
'''
Parse .poc file from CASTP web server. Lines are PDB format with pocket number at end:
ATOM 165 NH2 ARG A 45 -9.395 -35.779 0.675 1.00 32.68 2 POC
ATOM 194 O VAL B 27 -12.428 -9.493 5.152 1.00 13.76 1 POC
ATOM 196 CG1 VAL B 27 -11.788 -7.554 2.677 1.00 14.96 1 POC
'''
f = open(poc_file_path, 'r')
lines = f.readlines()
f.close()
pockets = {}
for line in lines:
if line.startswith('ATOM '):
atom_name, chain_id, res_num = line[12:16].strip(), line[21:22], int(line[22:26])
spec = '/%s:%d@%s' % (chain_id, res_num, atom_name)
pocket_num = int(line[66:71])
if pocket_num in pockets:
pockets[pocket_num].append(spec)
else:
pockets[pocket_num] = [spec]
pocket_specs = {pnum: ''.join(pspecs) for pnum, pspecs in pockets.items()}
return pocket_specs
def name_pockets(session, pocket_specs):
'''
Use ChimeraX name command to name the atoms for each pocket
pocket1, pocket2, ....
'''
from chimerax.basic_actions.cmd import name
for pnum, atoms_spec in pocket_specs.items():
name(session, 'pocket%d' % pnum, atoms_spec)
def color_pockets(session, n):
'''
Color n pockets randomly using named atoms pocket1, pocket2, ....
'''
from chimerax.core.colors import random_colors, hex_color
from chimerax.core.commands import run
for p, color in enumerate(random_colors(n)):
run(session, 'color pocket%d %s' % (p+1, hex_color(color)))
def readcastp(session, poc_file, color = False):
pocket_specs = pocket_atom_specifiers(poc_file)
name_pockets(session, pocket_specs)
if color:
color_pockets(session, len(pocket_specs))
print('%d pockets' % len(pocket_specs))
def register_command(logger):
from chimerax.core.commands import CmdDesc, register, OpenFileNameArg, BoolArg
desc = CmdDesc(
required = [('poc_file', OpenFileNameArg)],
keyword = [('color', BoolArg)],
synopsis = 'Read CASTP pockets file'
)
register('readcastp', desc, readcastp, logger=logger)
register_command(session.logger)
Tom Goddard, January 18, 2021