ChimeraX Recipes
Flip Atom Z Coordinates
Here is Python code defining a command flip that changes atom z coordinates z -> -z. This changes the hand of the atomic structure, for instance changing all L amino acids to D amino acids. Open flip.py in ChimeraX 1.0 to define the flip command
open flip.py
then use the command on the atomic model.
flip #1
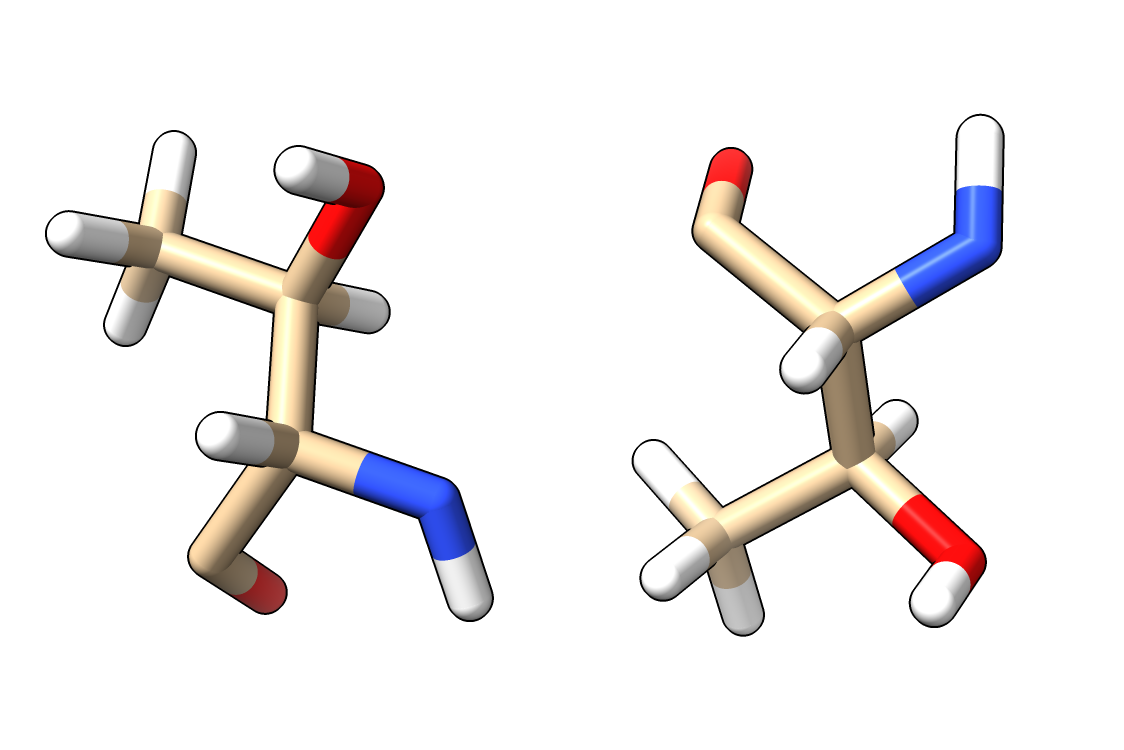
Here is the flip.py code:
# Create command to change atom coordiates z -> -z.
# Opening this file in ChimeraX 1.0 defines the flip command.
#
# flip #1
def flip(session, atoms):
xyz = atoms.coords
xyz[:,2] *= -1
atoms.coords = xyz
def register_command(session):
from chimerax.core.commands import CmdDesc, register
from chimerax.atomic import AtomsArg
desc = CmdDesc(required=[('atoms', AtomsArg)],
synopsis='flip atom z coordinates')
register('flip', desc, flip, logger=session.logger)
register_command(session)
Tom Goddard, September 3, 2020