ChimeraX Recipes
Plot ModelCIF pairwise residue scores
Here is Python code defining a command “modelcif pae” that plots pairwise residue scores found in the ma_qa_metric_local_pairwise table found in some ModelCIF files. These scores are often predicted aligned error values from AlphaFold. The command writes a file in AlhpaFold JSON PAE format and opens it with the alphafold pae command. Displaying pairwise scores was requested by Gerardo Tauriello on the ChimeraX mailing list.
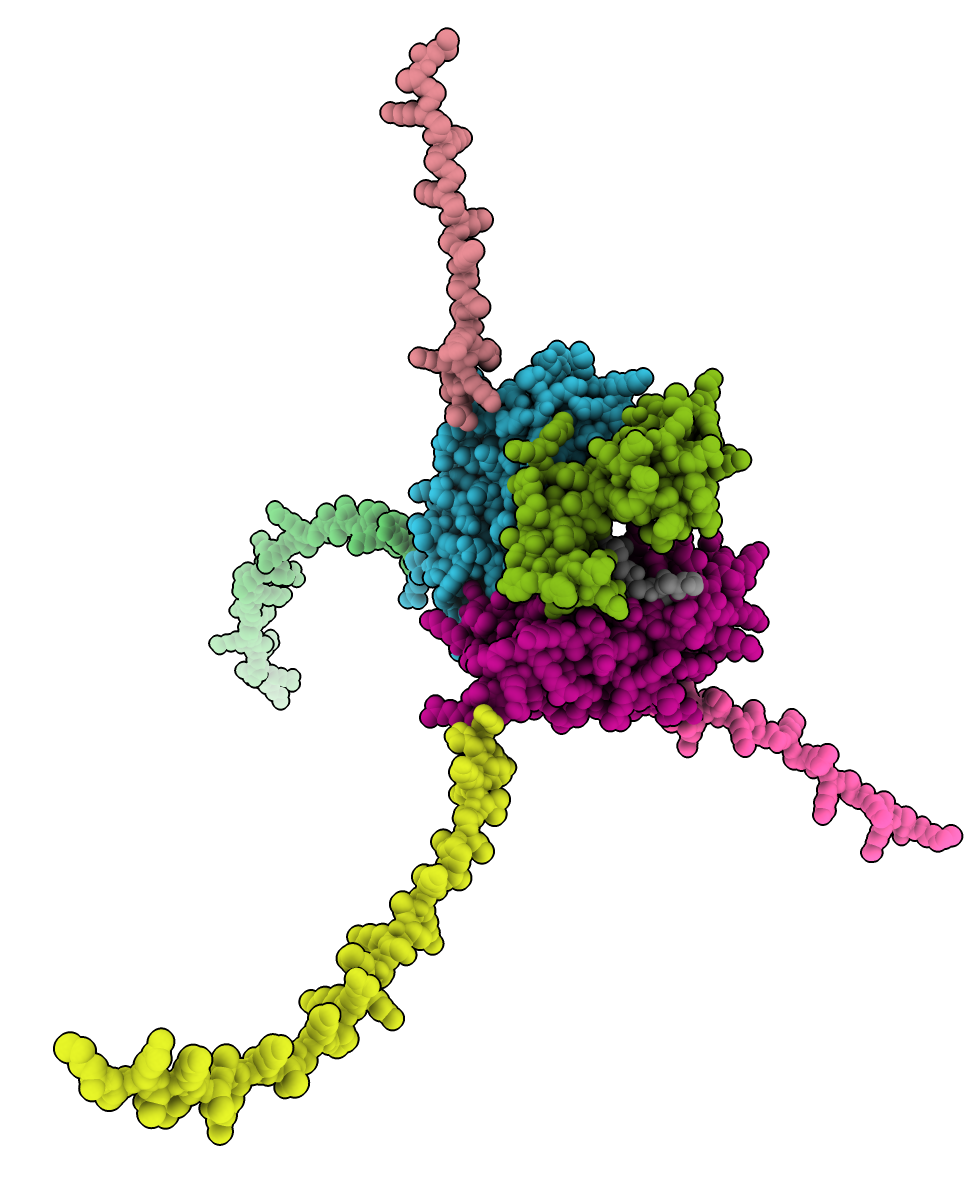
Here’s an ChimeraX example using a Pombe histone H3/H4 and DPB3 complex ModelCIF file 003-Spombe_H3-H4_tetramer_DPB3.cif.
open ~/Downloads/003-Spombe_H3-H4_tetramer_DPB3.cif
Open the modelcif_pae.py Python code to define the modelcif pae command
open ~/Downloads/modelcif_pae.py
Then create the score plot with
modelcif pae #1
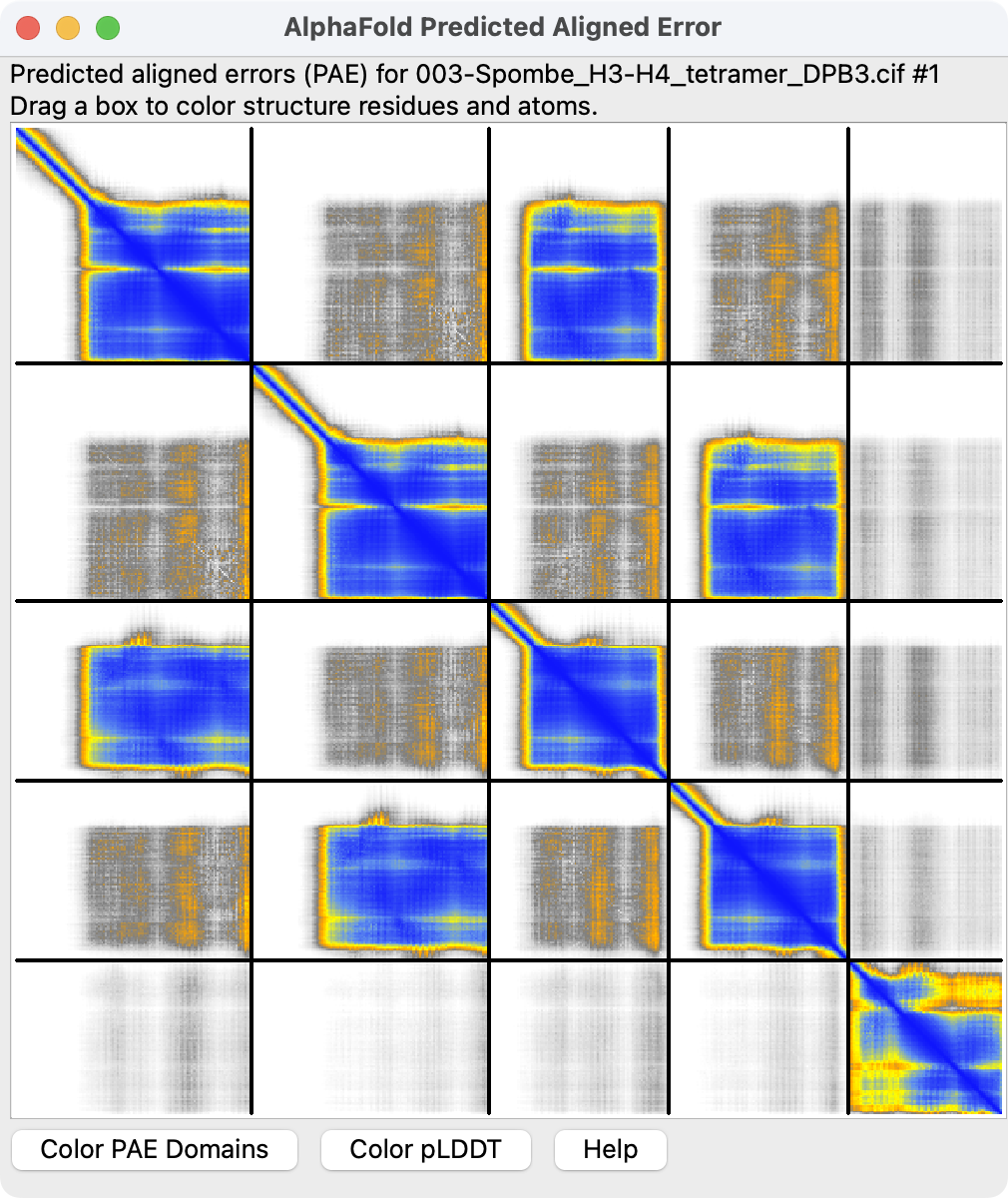
Here is the modelcif_pae.py code:
# Read pairwise residue scores from a ModelCIF file and plot them in ChimeraX.
def modelcif_pae(session, structure, json_output_path = None, metric_id = None, default_score = 100):
matrix = read_pairwise_scores(structure, metric_id = metric_id, default_score = default_score)
if json_output_path is None:
import tempfile
temp = tempfile.NamedTemporaryFile(prefix = 'modelcif_pae_', suffix = '.json')
json_output_path = temp.name
write_json_pae_file(json_output_path, matrix)
# Open PAE plot
from chimerax.core.commands import run, quote_if_necessary
open_cmd = f'alphafold pae #{structure.id_string} file {quote_if_necessary(json_output_path)}'
run(session, open_cmd)
def read_pairwise_scores(structure, metric_id = None, default_score = 100):
if not hasattr(structure, 'filename'):
from chimerax.core.errors import UserError
raise UserError(f'Structure {structure} has no associated file')
values = read_ma_qa_metric_local_pairwise_table(structure.filename)
if values is None:
from chimerax.core.errors import UserError
raise UserError(f'Structure file {structure.filename} contains no pairwise residue scores (i.e. no table "ma_qa_metric_local_pairwise")')
# Use only the scores with the given metric id.
if metric_id is None and len(values) > 0:
metric_id = values[0][5]
values = [v for v in values if v[5] == metric_id]
if len(values) == 0:
from chimerax.core.errors import UserError
raise UserError(f'Structure file {structure.filename} has no scores for metric id "{metric_id}"')
matrix_index = {(r.chain_id,r.number):ri for ri,r in enumerate(structure.residues)}
nr = structure.num_residues
from numpy import empty, float32
matrix = empty((nr,nr), float32)
matrix[:] = default_score
for model_id, chain_id_1, res_num_1, chain_id_2, res_num_2, metric_id, metric_value in values:
res_num_1, res_num_2, metric_value = int(res_num_1), int(res_num_2), float(metric_value)
r1 = matrix_index[(chain_id_1, res_num_1)]
r2 = matrix_index[(chain_id_2, res_num_2)]
matrix[r1,r2] = metric_value
return matrix
def read_ma_qa_metric_local_pairwise_table(path):
from chimerax.mmcif import get_cif_tables
table_names = ['ma_qa_metric_local_pairwise']
try:
tables = get_cif_tables(path, table_names)
except TypeError:
# Bug in get_cif_tables() results in TypeError if table not present. Ticket #16054
return None
if len(tables) == 0:
return None
ma_qa_metric_local_pairwise = tables[0]
field_names = ['model_id',
'label_asym_id_1', 'label_seq_id_1',
'label_asym_id_2', 'label_seq_id_2',
'metric_id', 'metric_value']
values = ma_qa_metric_local_pairwise.fields(field_names)
return values
def write_json_pae_file(json_output_path, matrix):
# Write matrix in JSON AlphaFold PAE format
# {"pae": [[17.14, 18.75, 17.91, ...], [5.32, 8.23, ...], ... ]}
n = matrix.shape[0]
dists = ', '.join(('[ ' + ', '.join('%.2f' % matrix[i,j] for j in range(n)) + ' ]')
for i in range(n))
with open(json_output_path, 'w') as file:
file.write('{"pae": [')
file.write(dists)
file.write(']}')
def register_command(logger):
from chimerax.core.commands import CmdDesc, register, StringArg, FloatArg, SaveFileNameArg
from chimerax.atomic import StructureArg
desc = CmdDesc(
required = [('structure', StructureArg)],
keyword = [('metric_id', StringArg),
('default_score', FloatArg),
('json_output_path', SaveFileNameArg)],
synopsis = 'Plot ModelCIF pairwise residue scores'
)
register('modelcif pae', desc, modelcif_pae, logger=logger)
register_command(session.logger)
Tom Goddard, October 1, 2024