ChimeraX Recipes
Select atoms inside map bounds
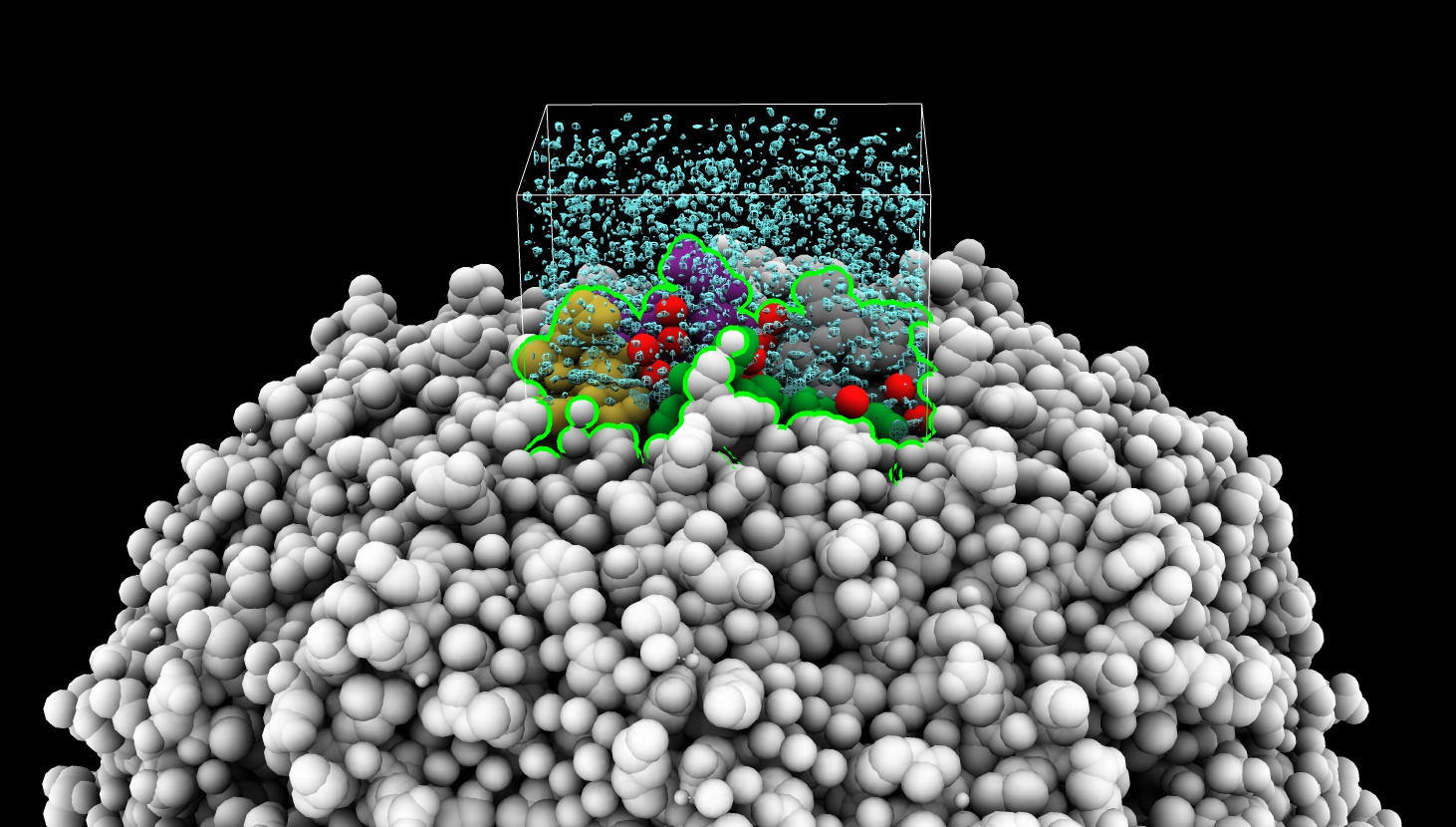
Here is a way to select atoms within a displayed map subregion using a command selectbox defined by the Python code below.
open 7k3v
open 22657 from emdb
volume #2 region 184,164,261,246,247,396
open selectbox.py
selectbox #1 inMap #2
Here is the select_box.py code that defines the selectbox command:
#
# Implement selectbox command that selects atoms within the bounds of shown map subregion.
#
# open select_box.py
# selectbox #1 in #2
#
def select_box(session, atoms, in_map):
# First convert atom coordinates to map coordinates in case
# one has been moved relative to the other.
pos = in_map.position.inverse().transform_points(atoms.scene_coords)
x,y,z = pos[:,0], pos[:,1], pos[:,2]
# Find atoms with coordinates in map bounds
(xmin,ymin,zmin), (xmax,ymax,zmax) = in_map.xyz_bounds()
mask = (x >= xmin) & (x <= xmax) & (y >= ymin) & (y <= ymax) & (z >= zmin) & (z <= zmax)
inside_atoms = atoms[mask]
# Select inside atoms
session.selection.clear()
inside_atoms.selected = True
inside_atoms.intra_bonds.selected = True
# Log a message saying how many atoms selected.
session.logger.status('Selected %d atoms in map %s bounds'
% (len(inside_atoms), in_map.name_with_id()),
log = True)
return inside_atoms
def register_selectbox_command(logger):
from chimerax.core.commands import CmdDesc, register
from chimerax.atomic import AtomsArg
from chimerax.map import MapArg
desc = CmdDesc(
required = [('atoms', AtomsArg)],
keyword = [('in_map', MapArg)],
required_arguments = ['in_map'],
synopsis = 'Select atoms in shown map subregion'
)
register('selectbox', desc, select_box, logger=logger)
register_selectbox_command(session.logger)
Tom Goddard, May 21, 2021