ChimeraX Recipes
Surface Exposed Residues
Here is how to select and color surface exposed residues that are hydrophobic.
Use measure sasa to define an “area” attribute of residues, then assign residue hydrophobicities from a file with the defattr command, and then use a single select command to intersect these two attributes.
In the defattr example files I already happened to include a file kdHydrophobicity.txt to assign Kyte-Doolittle hydrophobity, as well as for several other hydrophobicity scales. Just download whichever one(s) you want.
The commands could be something like:
open 6g2j
measure sasa protein
defattr ~/Desktop/kdHydrophobicity.txt
select ::area>25 & ::kdHydrophobicity>0.0
surface protein enclose protein
color sel magenta
… which looks like this …
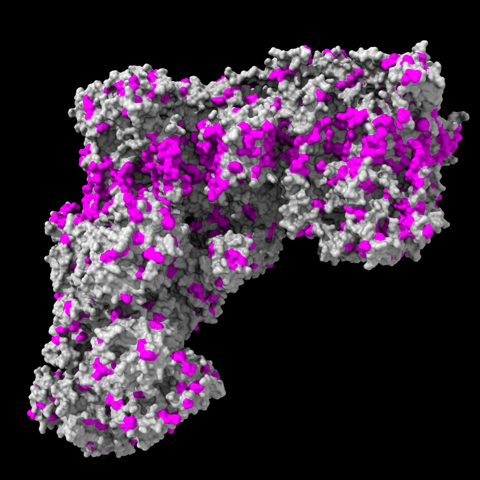
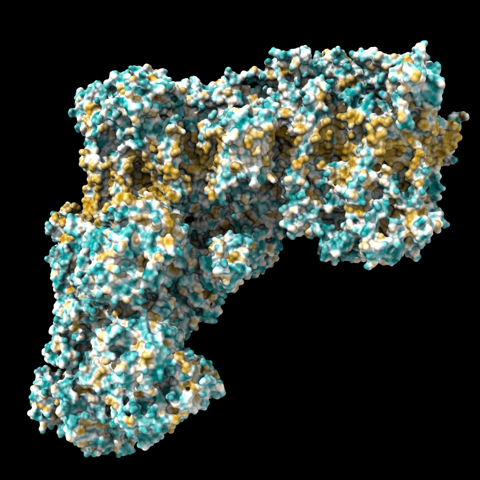
… in pretty good agreement with MLP coloring, e.g.
mlp surfaces #1.3
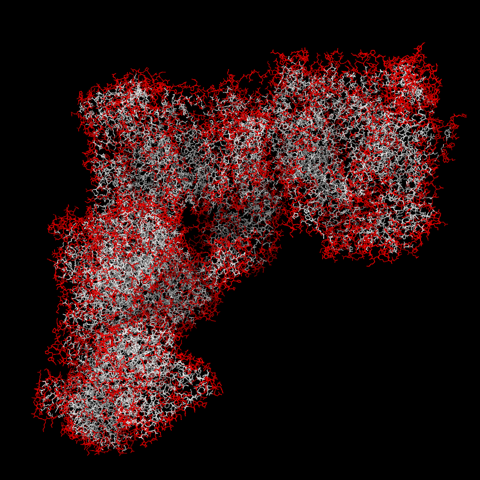
In the select command you may want to experiment with the surface-area and hydrophobicity cutoffs, as well as which hydrophobicity scale you use. I didn’t try a lot of values for surface-area cutoff, but here is with all residues with values >25 colored red, reasonable at least as a first approximation:
Elaine Meng, September 7, 2020